|
Below are some of the images that illustrate the work of our lab. Click on an image to get a larger picture.

The image on the left illustrates a graphical alignment of a DNA inverted repeat found with IRF and displayed within our web interface. The blue line represents the sequence going from left to right, and the red line represents the reverse complement running the opposite direction. If the pair matches (A-T, or G-C pair) a red empty square is displayed, otherwise the mismatch is dispayed . [ Large Image ]
| | |

This image is a multiple alignment of a family of tandem repeats found with TRDB. After repeats with similar pattern length were extracted, clustering within the resulting set, using the Biconnected Components Algorithm, detected families of similar repeats. The top blue line shows the pattern of the repeat which was used as the master (other repeats were aligned to it.) The Aqua lines represent patterns of other repeats. Red lines simply correspond to matching characters inside repeats. [ Large Image ]
| | |

Repeats as they are displayed and filtered in TRDB. The filters allow you to refine a set of repeats to those of particular interest. For example, in the picture, all repeats of pattern size longer than 60 nucleotides were removed and then some were selected manually with special checkboxes. Also, only those repeats overlapping or within 100 bp of a known or predicted gene were retained. Once the set is filtered, it is possible to save it as a new set. [ Large Image ]
| | |

This image displayes a histogram of tandem repeat pattern size in one chromosome of C. elegans as produced by TRDB. Distributions on many repeat characteristics are possible. Histograms can also be displayed in tabular form. [ Large Image ]
| | |

Another important feature of TRDB is the ability to modify which data are presented. Information available includes sequence characteristics of the repeats, polymorphism prediction and gene annotations. [ Large Image ]
| | |

Mutation master accepts a multiple alignment data file (MSF) and rapidly provides a visual display and tabulation of site, frequency, number and likelihood of point mutations. Image on the left displays a graph of the BLOSUM substitution score for frequently observed point mutations relative to a consensus sequence. [ Large Image ]
| | |
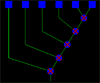
In TRDB, Sets are often created, filtered and merged. Set History page remembers all the modification that were done to the set during its lifetime. On the top of the page there is a description of the set. On the bottom of the page there is an image with a tree like structure. It shows you the history of the set. Blue boxes indicate sequences, green indicate filters and blue/red circles indicate merges. If the user clicks on one of the tree items, item information is extracted into the text area above. [ Large Image ]
| | |
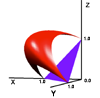
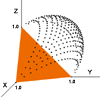
Clustering algorithms used by TRDB are based on a distance measure between repeats. One experimental measure uses the distance between points along a multidimensional entropy surface. Points on the surface represent the composition (A,C,G,T content) of a column in the alignment of a repeat. Entropy Surface is a program we wrote to generate the surfaces and create distance tables. A three dimensional entropy surface is displayed on the right. [ Large Image ] Shortest distance between points is found using Dykstra's algorithm on a graph of random points generated on the surface. Image on the left is a three dimensional visualization of an array of evenly spaced points on the surface. [ Large Image ]
|
|